Note
Go to the end to download the full example code.
Benchmark of Radius Clustering using multiple datasets and comparison with custom MDS#
This example demonstrates how to implement a custom solver for the MDS problem and use it within the Radius Clustering framework. Plus, it compares the results of a naive implementation using the NetworkX library with the Radius Clustering implementation.
- The example includes:
Defining the custom MDS solver.
Defining datasets to test the clustering.
Applying Radius clustering on the datasets using the custom MDS solver.
Ensure this solution works.
Establish a benchmark procedure to compare the Radius clustering with a naive implementation using NetworkX.
- Comparing the results in terms of :
Execution time
Number of cluster found
Visualizing the benchmark results.
Visualizing the clustering results.
This example is useful for understanding how to implement a custom MDS solver and how to perform an advanced usage of the package.
# Author: Haenn Quentin
# SPDX-License-Identifier: MIT
Import necessary libraries#
Since this example is a benchmark, we need to import the necessary libraries to perform the benchmark, including NetworkX for the naive implementation, matplotlib for visualization, and sklearn for the datasets.
import networkx as nx
import numpy as np
import matplotlib.pyplot as plt
import time
import warnings
from sklearn.datasets import fetch_openml
from radius_clustering import RadiusClustering
from sklearn.metrics import pairwise_distances_argmin
warnings.filterwarnings("ignore", category=RuntimeWarning, module="sklearn")
Define a custom MDS solver#
We define a custom MDS solver that uses the NetworkX library to compute the MDS. Note the signature of the function is identical to the one used in the RadiusClustering class.
def custom_solver(n: int, edges: np.ndarray, nb_edges: int, random_state=None):
"""
Custom MDS solver using NetworkX to compute the MDS problem.
Parameters:
-----------
n : int
The number of points in the dataset.
edges : np.ndarray
The edges of the graph, flattened into a 1D array.
nb_edges : int
The number of edges in the graph.
random_state : int | None
The random state to use for reproducibility.
Returns:
--------
centers : list
A sorted list of the centers of the clusters.
mds_exec_time : float
The execution time of the MDS algorithm in seconds.
"""
G = nx.Graph()
G.add_edges_from(edges)
start_time = time.time()
centers = list(nx.algorithms.dominating.dominating_set(G))
mds_exec_time = time.time() - start_time
centers = sorted(centers)
return centers, mds_exec_time
Define datasets to test the clustering#
We will use 4 datasets to test the clustering: 1. Iris dataset 2. Wine dataset 3. Breast Cancer dataset (WDBC) 4. Vehicle dataset These are common datasets used in machine learning and lead to pretty fast results. Structure of the variable DATASETS: - The key is the name of the dataset. - The value is a tuple containing:
The dataset fetched from OpenML.
The radius to use for the Radius clustering. (determined in literature, see references on home page)
DATASETS = {
"iris": (fetch_openml(name="iris", version=1, as_frame=False), 1.43),
"wine": (fetch_openml(name="wine", version=1, as_frame=False), 232.09),
"glass": (fetch_openml(name="glass", version=1, as_frame=False), 3.94),
"ionosphere": (fetch_openml(name="ionosphere", version=1, as_frame=False), 5.46),
"breast_cancer": (fetch_openml(name="wdbc", version=1, as_frame=False), 1197.42),
"synthetic": (fetch_openml(name="synthetic_control", version=1, as_frame=False), 70.12),
"vehicle": (fetch_openml(name="vehicle", version=1, as_frame=False), 155.05),
"yeast": (fetch_openml(name="yeast", version=1, as_frame=False), 0.4235),
}
Define the benchmark procedure#
We define a function to perform the benchmark on the datasets. The procedure is as follows: 1. Creates an instance of RadiusClustering for each solver. 2. For each instance, fit the algorithm on each dataset. 3. Store the execution time and the number of clusters found for each dataset. 4. Return the results as a dictionary.
def benchmark_radius_clustering():
results = {}
exact = RadiusClustering(manner="exact", radius=1.43)
approx = RadiusClustering(manner="approx", radius=1.43)
custom = RadiusClustering(
manner="custom", radius=1.43
)
custom.set_solver(custom_solver) # Set the custom solver
algorithms = [exact, approx, custom]
# Loop through each algorithm and dataset
for algo in algorithms:
algo_results = {}
time_algo = []
clusters_algo = []
# Loop through each dataset
for name, (dataset, radius) in DATASETS.items():
X = dataset.data
# set the radius for the dataset considered
setattr(algo, "radius", radius)
# Fit the algorithm
t0 = time.time()
algo.fit(X)
t_algo = time.time() - t0
# Store the results
time_algo.append(t_algo)
clusters_algo.append(len(algo.centers_))
algo_results["time"] = time_algo
algo_results["clusters"] = clusters_algo
results[algo.manner] = algo_results
return results
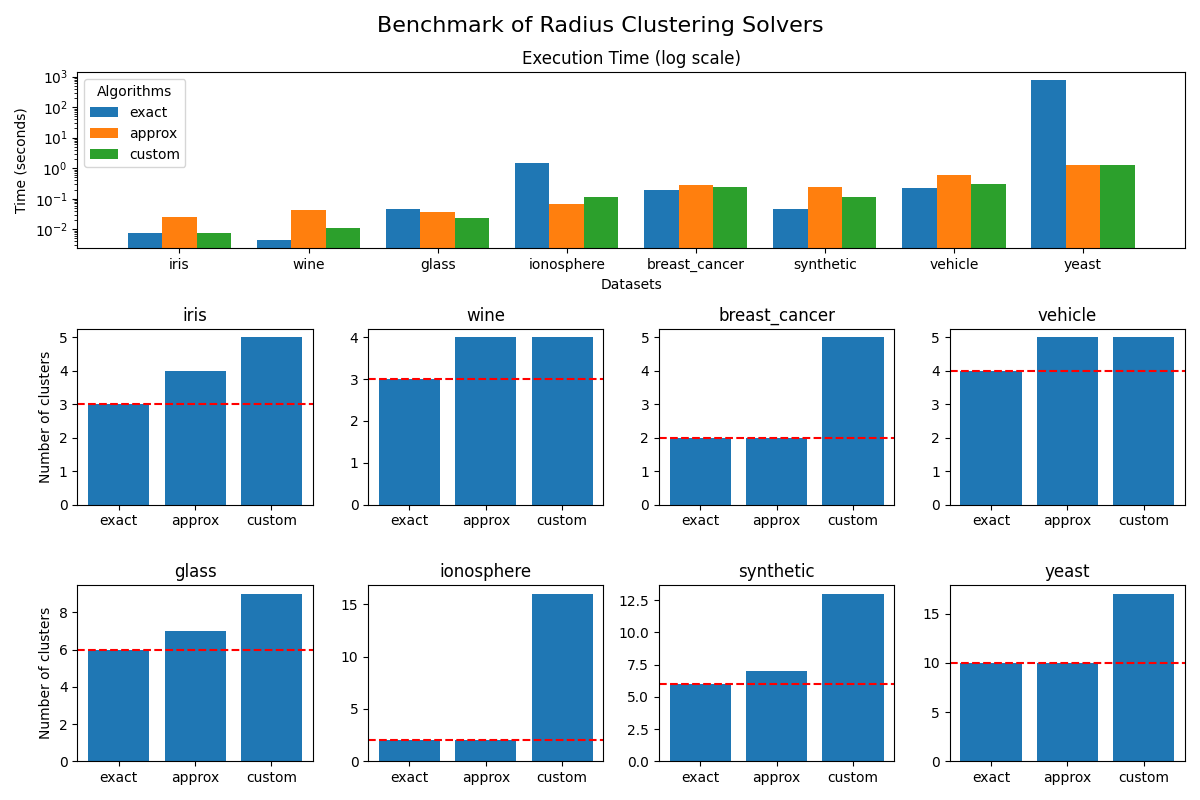
Run the benchmark and plot the results#
We run the benchmark and plot the results for each dataset.
results = benchmark_radius_clustering()
# Plot the results
fig, axs = plt.subplot_mosaic(
[
["time", "time", "time", "time"],
["iris", "wine", "breast_cancer", "vehicle"],
["glass", "ionosphere", "synthetic", "yeast"],
],
layout="constrained",
figsize=(12, 8),
)
fig.suptitle("Benchmark of Radius Clustering Solvers", fontsize=16)
axs['time'].set_yscale('log') # Use logarithmic scale for better visibility
algorithms = list(results.keys())
dataset_names = list(DATASETS.keys())
n_algos = len(algorithms)
x_indices = np.arange(len(dataset_names)) # the label locations
bar_width = 0.8 / n_algos # the width of the bars, with some padding
for i, algo in enumerate(algorithms):
times = results[algo]["time"]
# Calculate position for each bar in the group to center them
position = x_indices - (n_algos * bar_width / 2) + (i * bar_width) + bar_width / 2
axs['time'].bar(position, times, bar_width, label=algo)
for i, (name, (dataset, _)) in enumerate(DATASETS.items()):
axs[name].bar(
results.keys(),
[results[algo]["clusters"][i] for algo in results.keys()],
label=name,
)
axs[name].axhline(
y=len(set(dataset.target)), # Number of unique classes in the dataset
label="True number of clusters",
color='r',
linestyle='--',
)
axs[name].set_title(name)
axs["iris"].set_ylabel("Number of clusters")
axs["glass"].set_ylabel("Number of clusters")
axs['time'].set_title("Execution Time (log scale)")
axs['time'].set_xlabel("Datasets")
axs['time'].set_ylabel("Time (seconds)")
axs['time'].set_xticks(x_indices)
axs['time'].set_xticklabels(dataset_names)
axs['time'].legend(title="Algorithms")
plt.tight_layout()
plt.show()
/home/runner/work/radius_clustering/radius_clustering/examples/plot_benchmark_custom.py:223: UserWarning: The figure layout has changed to tight
plt.tight_layout()
Conclusion#
In this example, we applied Radius clustering to the Iris and Wine datasets and compared it with KMeans clustering. We visualized the clustering results and the difference between the two clustering algorithms. We saw that Radius Clustering can lead to smaller clusters than kmeans, which produces much more equilibrate clusters. The difference plot can be very useful to see where the two clustering algorithms differ.
Total running time of the script: (13 minutes 59.426 seconds)
Related examples